A team led by the Hong Kong University of Science and Technology (HKUST) has developed a technique to study how different fish species interact with each other in a coastal region, a breakthrough that helps explain the complex relationships among marine species and how global warming impacts fish populations.
By analyzing minute traces of fish DNA from samples of seawater, the team combined the use of environmental DNA – known as eDNA – and advanced statistical analysis to not only detect the presence of fish species, but also reveal how the species interact with each other.
The use of eDNA to monitor biodiversity has surged in popularity in recent years, particularly for detecting aquatic organisms like fish. As animals move through their environment, they shed fragments of genetic material, such as skin cells, waste products, and other body fluids. By extracting these traces of DNA from samples of water, soil, or air, scientists can determine the presence and diversity of species with high accuracy.
But previous eDNA studies have mostly been limited to detecting the presence or absence of certain species. To get a better understanding and monitoring of ecosystems, it is necessary to estimate the quantity of fish species and detect the interspecific interactions, or interactions among species.
The team, led by Prof. Masayuki USHIO, Assistant Professor of the Department of Ocean Science at HKUST and Dr. Masaki MIYA at Natural History Museum and Institute, Chiba, Japan, developed a new technique to achieve the abovementioned purpose by analyzing high-frequency time-series data from fish eDNA, making it possible to comprehensively monitor interactions among species.
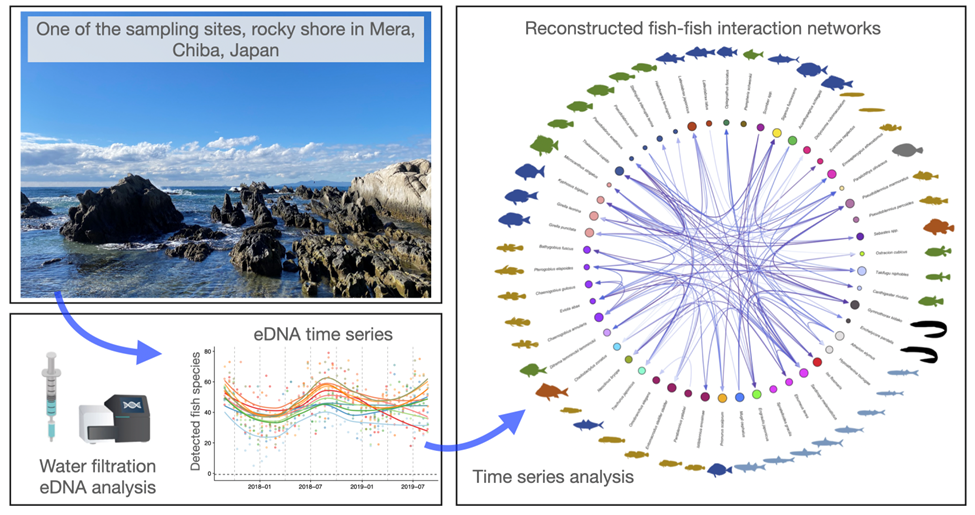
Interspecific interactions, such as prey-predator, competitive, and mutualistic relations, have a significant impact on ecosystem dynamics. The research paves the way for scientists to contribute to more accurate assessment of the ecosystem status and future predictions of its dynamics.
The researchers set 11 study sites along the coast of the Boso Peninsula in Chiba Prefecture, Japan, where they conducted biweekly water sampling for two years. They extracted eDNA from the collected samples and analyzed fish communities using a method which generated the eDNA-based high-frequency time series of fish communities in coastal ecosystems. Then, they detected and quantified the strength of fish-fish interspecific interactions in the communities by analyzing the high-frequency eDNA time-series data using cutting-edge time-series analysis methods.
Among their important findings was that water temperature can have both positive and negative effects on how different fish species interact with each other. They also found that different fish species can react differently to changes in temperature, shedding light on how global warming can impact the complex relationships between fish species in coastal areas.
The findings were recently published in international academic journal eLife.

“By integrating state-of-the-art techniques from different scientific fields, we showed that eDNA analysis can estimate not only ‘what’ and ‘how many’ species are present, but also ‘who interacts with whom’”, Prof. Ushio says.
The framework can be used to study interactions between different types of organisms, not just fish. This includes microbes, crustaceans, and other invertebrates. For example, scientists could use the technique to study how different organisms interact in aquaculture systems. It could also help identify potential harmful pathogens that could impact commercially important fish species.
In the long run, he said the research could help scientists and policymakers better understand how climate change is impacting fish populations, including commercially important and rare species.

“This information can be used to develop better conservation strategies to protect these species and ensure the long-term sustainability of oceans. For example, to conserve a rare species, it may be important to also protect a fish species that has a significant impact on the rare species,” he says.
The team will soon explore new technologies, such as automated water sampling systems and seawater sampling with underwater drones, to make eDNA analyses more efficient and effective, to further our understanding of how organisms interact with each other in nature, such as how pathogens impact fish populations.
They also plan to use advanced DNA sequencing technology to investigate eDNA sequences in more detail, such as with long-read sequencing.
“By utilizing these new technologies and collaborating with experts from different fields, we can continue to advance our understanding of how climate change impacts marine ecosystems, and develop better ways to protect them,” he says.









