A research team co-led by a scientist at the Hong Kong University of Science and Technology (HKUST) has developed a method to study how HIV mutates to escape the immune system in multiple patients, which could inform HIV vaccine design.
HIV, which can lead to AIDS, evolves rapidly and attacks the body’s immune system. Genetic mutations in the virus enable it to evade immune responses mounted by T cells and antibodies, which makes it all the more difficult to design an effective solution. While there is no effective cure for the virus currently available, it can be controlled with medication.
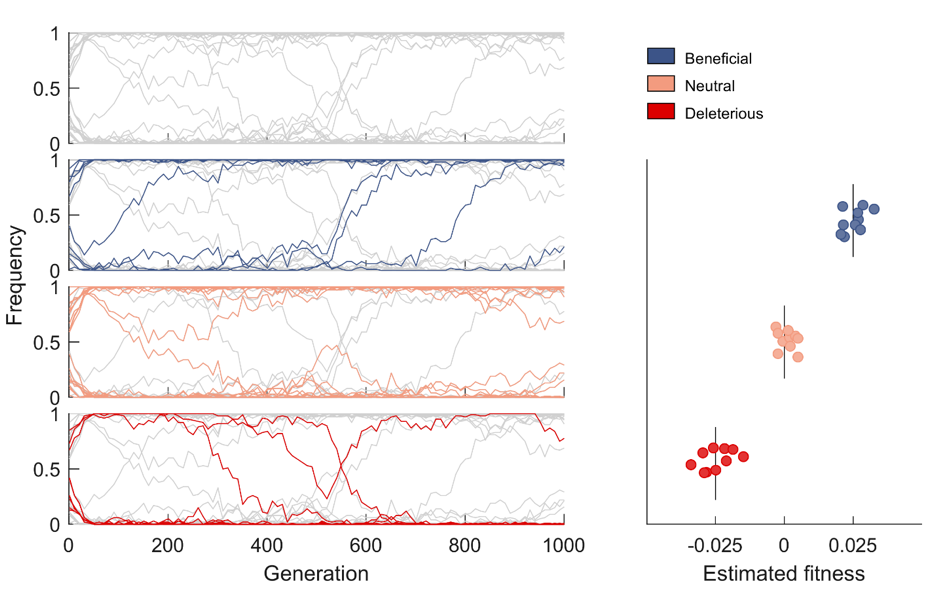
Now, the international research team has devised a new method from conventional statistical physics to reveal patterns of selection in HIV evolution using 14 patient data sets, providing a means to efficiently distinguishing the mutations that help the virus escape the immune system from those that are only random variations.
“Our novel method enables us to sort out which genetic changes provide an evolutionary advantage from those that offer no advantage or have a deleterious effect,” said Prof. Matthew MCKAY, a Professor from the Departments of Electronic and Computer Engineering and Chemical and Biological Engineering, who co-led the study with Prof. John BARTON, an Assistant Professor of Physics and Astronomy at University of California, Riverside.
“The method is quite general and could be used to study diverse evolutionary processes, such as the evolution of drug resistance of pathogens and the evolution of cancers. The accuracy and high efficiency of our approach enable the analysis of selection in complex evolutionary systems that were beyond the reach of existing methods,” he added.
“Understanding the genetic drivers of disease is important in the biomedical sciences,” said Prof. Barton. “Being able to identify genomic rearrangements is key to understanding how illnesses occur and how to treat them.” Notable examples of genetic drivers of disease include mutations that allow viruses to escape from immune control, while others confer drug resistance to bacteria.
“It can be difficult, however, to differentiate between real, adaptive mutations and random genetic variation,” he added. “The new method we developed allows us to identify such mutations in complex evolving populations.”
“However, it is computationally difficult to extract this information from data,” he said. “We used methods from statistical physics to overcome this computational challenge. Our method can be applied generally to evolving populations and is not limited to HIV.”
Some well-known diseases that have known genetic causes are cystic fibrosis, sickle cell anemia, Duchenne muscular dystrophy, colorblindness, and Huntington’s disease.
“Our approach isn't limited to HIV, but there are a few reasons why we focused on HIV as a test system,” Prof. McKay said. “HIV has an extraordinary capability to mutate within humans to escape from the immune system. However, the details of these immune escape dynamics are not well understood. If we can gain a clearer picture of how HIV evolves within a person, this may help to develop better treatments against the virus.”
Their findings were just published in high impact scientific journal Nature Biotechnology on November 30, 2020.
The research team also consists of Post-doctoral Fellow Dr. Muhammad Saqib SOHAIL of HKUST and Post-doctoral Fellow Dr. Raymond LOUIE of the Kirby Institute, University of New South Wales in Australia, who were joint first authors on the paper.